Pitt virologists and computational biologists are combining their expertise to understand the replication cycle of a deadly mosquito-borne alphavirus.
Eastern equine encephalitis virus, or EEEV, causes inflammation of the brain that can lead to confusion, seizures and loss of consciousness. It also has a high human mortality rate— about 30-70% of symptomatic cases are fatal. Just last year, at least seven states in the Northeast had confirmed cases of EEEV.
“One of the reasons the human mortality rate is so high is by the time people come to the hospital, the virus has already entered the central nervous system, and they have encephalitis when they show up,” said William Klimstra, an associate professor in the Department of Immunology.
EEEV has a short treatment window, and there are currently no licensed, targeted therapies for the virus. New treatments that can fit within the window of diagnosis are needed. To address this problem, Klimstra partnered with computational biologists James Faeder and Caroline Larkin to create a kinetic model of EEEV.
Faeder, an associate professor in the Department of Computational and Systems Biology, specializes in developing mathematical models of biological regulatory processes. He became interested in modeling viral replication within cells as a result of the COVID-19 pandemic. Before this point, there were few computational models depicting the biochemical detail of viral replication.
“If you build a model that doesn’t explicitly show the places where the drug molecules act on the cell and the virus, then you’re approximating what a drug is doing, as opposed to modeling in a specific way what it’s doing,” Faeder said.
By creating a model of EEEV, the research team’s goal was to look for vulnerabilities based on predictions of viral replication. If scientists can inhibit the points identified in the model, then this will play a key role in understanding EEEV and limiting the amount of virus produced.
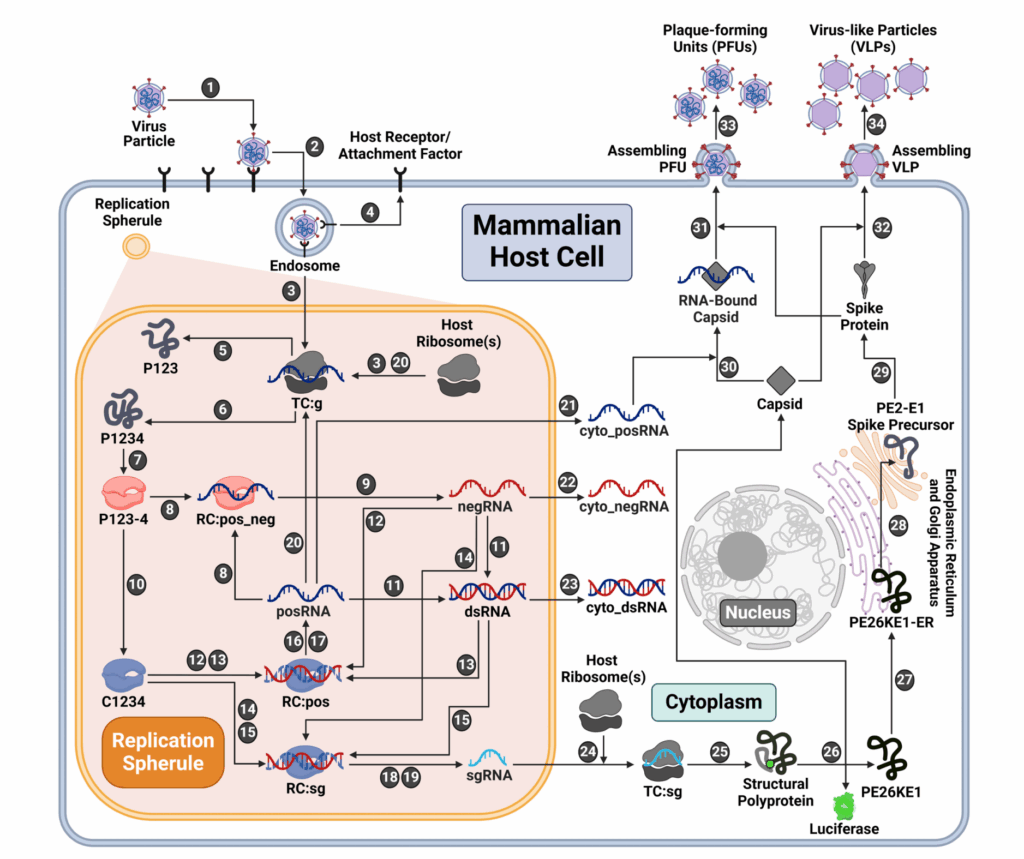
Larkin, a graduate of the Joint Carnegie Mellon–University of Pittsburgh PhD Program in Computational Biology, created the model. Building this model required her to strengthen her expertise in the field of virology in addition to computational biology. This led Larkin to her current postdoctoral position in the Department of Immunology.
“The model can simulate the dynamics for individual RNA strands, and that will provide further insight for William to do more experiments,” she said. “We can make different predictions about drug targeting and what effect it will have on viral production.”
The model made predictions about factors influencing virus replication within the cell. These different aspects of replication affect output, which is the number of virus particles produced within a cell.
Building the model is the first step in the process of understanding EEEV. The team has recently submitted a grant proposal to pursue a second step of subjecting the virus to innate immune responses.
“We created an incredibly detailed base for subsequently layering on different things that influence virus replication, like the cell’s lifespan and intracellular composition. And from this, we are hoping to identify vulnerabilities that drugs can then be used to treat,” Klimstra said.


